|
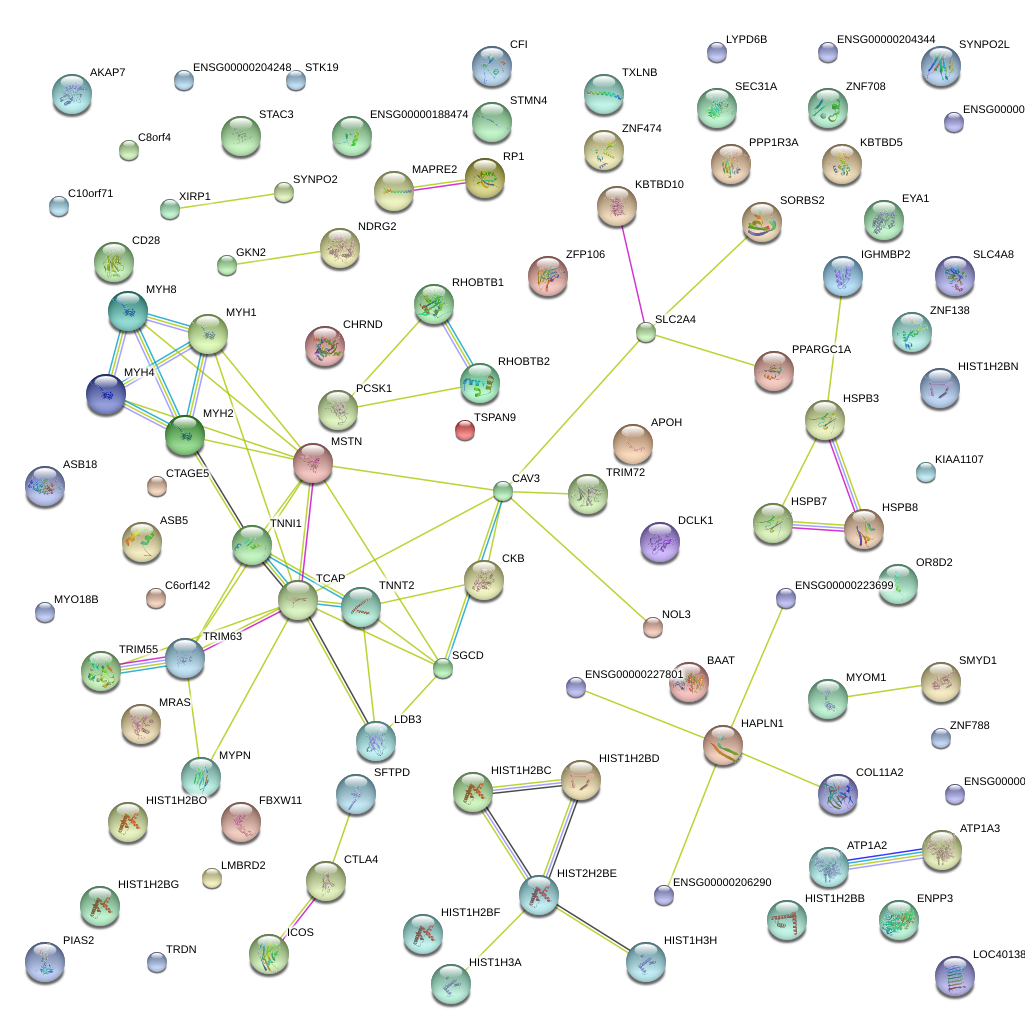
chr10_+_69539255
|
1.958
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr3_-_39209075
|
1.855
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr6_-_123999640
|
1.744
|
NM_006073
|
TRDN
|
triadin
|
|
chr5_+_155686333
|
1.633
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr2_+_88148413
|
1.551
|
NM_198274
|
SMYD1
|
SET and MYND domain containing 1
|
|
chr8_+_40130143
|
1.450
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr2_-_190635566
|
1.324
|
NM_005259
|
MSTN
|
myostatin
|
|
chr3_+_42702014
|
1.095
|
NM_152393
|
KBTBD5
|
kelch repeat and BTB (POZ) domain containing 5
|
|
chr18_-_3209986
|
1.086
|
NM_003803
NM_019856
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr6_+_27885692
|
1.063
|
NM_003536
|
HIST1H3H
|
histone cluster 1, H3h
|
|
chr2_+_204279442
|
1.060
|
NM_006139
|
CD28
|
CD28 molecule
|
|
chr7_-_113346284
|
1.049
|
NM_002711
|
PPP1R3A
|
protein phosphatase 1, regulatory (inhibitor) subunit 3A
|
|
chr5_-_83052606
|
1.021
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chr4_-_23500706
|
0.990
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr6_-_139654807
|
0.975
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr6_+_26266311
|
0.964
|
NM_021063
NM_138720
|
HIST1H2BD
|
histone cluster 1, H2bd
|
|
chr22_+_24468168
|
0.951
|
|
MYO18B
|
myosin XVIIIB
|
|
chr2_-_69033515
|
0.940
|
NM_182536
|
GKN2
|
gastrokine 2
|
|
chr10_-_62373828
|
0.931
|
NM_014836
|
RHOBTB1
|
Rho-related BTB domain containing 1
|
|
chr12_-_55931172
|
0.911
|
NM_145064
|
STAC3
|
SH3 and cysteine rich domain 3
|
|
chr17_+_7125702
|
0.901
|
NM_001042
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr17_+_35075124
|
0.895
|
NM_003673
|
TCAP
|
titin-cap (telethonin)
|
|
chr22_+_24468110
|
0.886
|
NM_032608
|
MYO18B
|
myosin XVIIIB
|
|
chr1_+_92405196
|
0.871
|
NM_015237
|
KIAA1107
|
KIAA1107
|
|
chr2_+_233099113
|
0.865
|
NM_000751
|
CHRND
|
cholinergic receptor, nicotinic, delta
|
|
chr14_+_38805622
|
0.844
|
|
CTAGE5
|
CTAGE family, member 5
|
|
chr3_+_8750485
|
0.827
|
NM_033337
NM_001234
|
CAV3
|
caveolin 3
|
|
chr1_-_199657468
|
0.824
|
NM_003281
|
TNNI1
|
troponin I type 1 (skeletal, slow)
|
|
chr10_-_75085754
|
0.796
|
NM_001114133
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr3_+_139550134
|
0.792
|
NM_001085049
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr10_+_50177192
|
0.773
|
NM_001135196
NM_199459
|
C10orf71
|
chromosome 10 open reading frame 71
|
|
chr6_-_26232109
|
0.770
|
NM_003526
|
HIST1H2BG
HIST1H2BC
|
histone cluster 1, H2bg
histone cluster 1, H2bc
|
|
chr1_-_16217025
|
0.749
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr4_-_186934059
|
0.735
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_-_33268177
|
0.731
|
NM_001163771
NM_080679
NM_080680
NM_080681
|
COL11A2
|
collagen, type XI, alpha 2
|
|
chr6_+_132000134
|
0.725
|
NM_005021
|
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3
|
|
chr17_-_61655915
|
0.722
|
NM_000042
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I)
|
|
chr10_-_81698840
|
0.692
|
NM_003019
|
SFTPD
|
surfactant protein D
|
|
chr2_-_236837726
|
0.683
|
NM_212556
|
ASB18
|
ankyrin repeat and SOCS box containing 18
|
|
chr1_-_199613427
|
0.681
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr9_-_103185621
|
0.678
|
NM_001127610
|
BAAT
|
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase)
|
|
chr17_-_10313600
|
0.675
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr16_+_31132842
|
0.668
|
NM_001008274
|
TRIM72
|
tripartite motif containing 72
|
|
chr5_+_53787187
|
0.657
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr6_+_131508153
|
0.654
|
NM_016377
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr13_-_35327798
|
0.650
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr17_-_10265991
|
0.640
|
NM_002472
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal
|
|
chr7_-_96471544
|
0.638
|
|
DLX6-AS1
|
DLX6 antisense RNA 1 (non-protein coding)
|
|
chr6_+_53991672
|
0.632
|
NM_138569
|
C6orf142
|
chromosome 6 open reading frame 142
|
|
chr2_+_204279624
|
0.629
|
|
CD28
|
CD28 molecule
|
|
chr2_+_170074457
|
0.618
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr5_+_121493113
|
0.603
|
NM_207317
|
ZNF474
|
zinc finger protein 474
|
|
chr8_+_55691179
|
0.601
|
NM_006269
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant)
|
|
chr1_+_556968
|
0.595
|
|
|
|
|
chr4_-_110942783
|
0.595
|
NM_000204
|
CFI
|
complement factor I
|
|
chr14_-_103058568
|
0.590
|
|
CKB
|
creatine kinase, brain
|
|
chr8_+_67201684
|
0.584
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr12_+_118100977
|
0.580
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr8_-_27171814
|
0.580
|
NM_030795
|
STMN4
|
stathmin-like 4
|
|
chr1_-_26266571
|
0.575
|
NM_032588
|
TRIM63
|
tripartite motif containing 63
|
|
chr6_+_131613196
|
0.574
|
NM_004842
NM_138633
|
AKAP7
|
A kinase (PRKA) anchor protein 7
|
|
chr18_-_3209846
|
0.571
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr4_+_120029426
|
0.565
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr7_+_63892198
|
0.544
|
NM_001160183
NM_006524
|
ZNF138
|
zinc finger protein 138
|
|
chr1_+_557269
|
0.544
|
|
|
|
|
chr5_-_95774444
|
0.541
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr7_-_91632525
|
0.540
|
NM_001161528
|
LOC401387
|
leucine-rich repeat and death domain-containing protein
|
|
chr19_+_12064077
|
0.536
|
|
ZNF788
|
zinc finger family member 788
|
|
chr8_-_72431274
|
0.535
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr4_-_177427252
|
0.532
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr11_-_123695302
|
0.532
|
NM_001002918
|
OR8D2
|
olfactory receptor, family 8, subfamily D, member 2
|
|
chr2_+_204509698
|
0.531
|
NM_012092
|
ICOS
|
inducible T-cell co-stimulator
|
|
chr10_+_88418147
|
0.525
|
NM_001171610
|
LDB3
|
LIM domain binding 3
|
|
chr2_+_149603226
|
0.516
|
NM_177964
|
LYPD6B
|
LY6/PLAUR domain containing 6B
|
|
chr5_-_36187710
|
0.503
|
NM_001007527
|
LMBRD2
|
LMBR1 domain containing 2
|
|
chr14_-_20563698
|
0.497
|
|
NDRG2
|
NDRG family member 2
|
|
chr14_-_20563722
|
0.496
|
NM_016250
NM_201535
NM_201536
NM_201537
|
NDRG2
|
NDRG family member 2
|
|
chr14_-_103058865
|
0.495
|
NM_001823
|
CKB
|
creatine kinase, brain
|
|
chr1_+_158352143
|
0.492
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr1_-_27113018
|
0.492
|
NM_021969
|
NR0B2
|
nuclear receptor subfamily 0, group B, member 2
|
|
chr1_-_150328153
|
0.491
|
NM_001008536
|
TCHHL1
|
trichohyalin-like 1
|
|
chr9_-_99740243
|
0.483
|
NM_197978
|
HEMGN
|
hemogen
|
|
chr12_+_100394787
|
0.483
|
NM_152323
|
SPIC
|
Spi-C transcription factor (Spi-1/PU.1 related)
|
|
chr2_-_199922047
|
0.482
|
|
SATB2
|
SATB homeobox 2
|
|
chr4_+_41057322
|
0.481
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr7_+_154943466
|
0.481
|
NM_001427
|
EN2
|
engrailed homeobox 2
|
|
chr2_+_238712101
|
0.481
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr4_+_88748704
|
0.480
|
NM_014208
|
DSPP
|
dentin sialophosphoprotein
|
|
chr9_-_205892
|
0.478
|
NM_152569
|
C9orf66
|
chromosome 9 open reading frame 66
|
|
chr9_-_14858974
|
0.477
|
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr1_+_65503011
|
0.476
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr17_-_10500189
|
0.475
|
NM_002470
|
MYH3
|
myosin, heavy chain 3, skeletal muscle, embryonic
|
|
chr11_+_1897374
|
0.474
|
NM_001042780
NM_001042781
NM_001042782
NM_006757
|
TNNT3
|
troponin T type 3 (skeletal, fast)
|
|
chr9_+_134884627
|
0.473
|
|
EEF1A1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr21_+_14510336
|
0.471
|
NM_144770
|
RBM11
|
RNA binding motif protein 11
|
|
chr4_-_110942576
|
0.466
|
|
CFI
|
complement factor I
|
|
chr3_+_139550368
|
0.464
|
|
MRAS
|
muscle RAS oncogene homolog
|
|
chr4_+_120276386
|
0.460
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr10_-_75080784
|
0.459
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr1_+_144124547
|
0.459
|
NM_145277
NM_202004
NM_213652
NM_213653
|
HFE2
|
hemochromatosis type 2 (juvenile)
|
|
chr1_-_181540631
|
0.454
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr7_-_150847698
|
0.450
|
|
RHEB
|
Ras homolog enriched in brain
|
|
chr18_+_18969724
|
0.448
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr21_-_18697829
|
0.447
|
NM_002772
|
TMPRSS15
|
transmembrane protease, serine 15
|
|
chr12_-_47679459
|
0.445
|
|
DDN
|
dendrin
|
|
chr11_-_101292462
|
0.444
|
NM_178127
|
ANGPTL5
|
angiopoietin-like 5
|
|
chr2_+_168383427
|
0.444
|
NM_020981
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr10_-_52315436
|
0.443
|
NM_001198818
NM_001198819
NM_001198820
NM_014576
NM_138932
NM_138933
|
A1CF
|
APOBEC1 complementation factor
|
|
chr3_-_150422474
|
0.439
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr10_+_88418243
|
0.439
|
NM_001080116
NM_001080114
NM_001080115
NM_001171611
NM_007078
|
LDB3
|
LIM domain binding 3
|
|
chr12_+_50731457
|
0.435
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr17_+_29606408
|
0.434
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr6_-_27914095
|
0.430
|
NM_003510
|
HIST1H2AK
|
histone cluster 1, H2ak
|
|
chr1_+_201729893
|
0.430
|
NM_014359
|
OPTC
|
opticin
|
|
chr3_+_185300660
|
0.429
|
NM_182589
|
HTR3E
|
5-hydroxytryptamine (serotonin) receptor 3, family member E
|
|
chr1_+_62835759
|
0.429
|
NM_014495
|
ANGPTL3
|
angiopoietin-like 3
|
|
chr7_-_150847624
|
0.428
|
|
RHEB
|
Ras homolog enriched in brain
|
|
chr14_+_99309688
|
0.428
|
|
EML1
|
echinoderm microtubule associated protein like 1
|
|
chr19_+_3884100
|
0.417
|
NM_170678
|
ITGB1BP3
|
integrin beta 1 binding protein 3
|
|
chr15_+_76171981
|
0.417
|
NM_001101404
|
SH2D7
|
SH2 domain containing 7
|
|
chr1_-_202595666
|
0.417
|
NM_014935
|
PLEKHA6
|
pleckstrin homology domain containing, family A member 6
|
|
chr21_-_33107813
|
0.415
|
NM_001162495
NM_001162496
NM_019596
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr1_-_23623713
|
0.414
|
NM_003196
|
TCEA3
|
transcription elongation factor A (SII), 3
|
|
chr4_-_64957595
|
0.413
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr7_-_135263737
|
0.411
|
|
MTPN
LUZP6
|
myotrophin
leucine zipper protein 6
|
|
chr17_-_31441390
|
0.411
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr16_-_70591377
|
0.411
|
NM_181536
|
PKD1L3
|
polycystic kidney disease 1-like 3
|
|
chr2_-_148495642
|
0.408
|
NM_181742
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr7_+_129693938
|
0.407
|
NM_001869
|
CPA2
|
carboxypeptidase A2 (pancreatic)
|
|
chr8_-_124818754
|
0.404
|
NM_001003954
NM_004306
|
ANXA13
|
annexin A13
|
|
chr13_-_50315832
|
0.404
|
NM_198989
|
DLEU7
|
deleted in lymphocytic leukemia, 7
|
|
chr14_-_69616616
|
0.403
|
NM_182936
NM_001130417
|
SLC8A3
|
solute carrier family 8 (sodium/calcium exchanger), member 3
|
|
chr4_+_74566325
|
0.403
|
NM_001133
|
AFM
|
afamin
|
|
chr9_-_96395828
|
0.402
|
NM_003837
|
FBP2
|
fructose-1,6-bisphosphatase 2
|
|
chr11_+_48466844
|
0.400
|
NM_001005512
|
OR4A47
|
olfactory receptor, family 4, subfamily A, member 47
|
|
chr6_-_128281434
|
0.398
|
NM_001164687
|
THEMIS
|
thymocyte selection associated
|
|
chr14_+_31868229
|
0.397
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr1_+_42773146
|
0.396
|
NM_001080850
|
CCDC30
|
coiled-coil domain containing 30
|
|
chr10_-_92671005
|
0.396
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr1_-_117554999
|
0.393
|
NM_024626
|
VTCN1
|
V-set domain containing T cell activation inhibitor 1
|
|
chr18_+_20294590
|
0.392
|
NM_001143828
NM_001160166
NM_021624
|
HRH4
|
histamine receptor H4
|
|
chr10_-_52315390
|
0.392
|
|
A1CF
|
APOBEC1 complementation factor
|
|
chr2_-_228206045
|
0.389
|
NM_001162483
NM_020161
|
C2orf83
|
chromosome 2 open reading frame 83
|
|
chr7_-_150847529
|
0.388
|
|
RHEB
|
Ras homolog enriched in brain
|
|
chr1_-_116184846
|
0.385
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr19_+_57540493
|
0.384
|
NM_173530
|
ZNF610
|
zinc finger protein 610
|
|
chr1_-_157014048
|
0.383
|
NM_001005278
|
OR6N2
|
olfactory receptor, family 6, subfamily N, member 2
|
|
chr10_-_69267774
|
0.382
|
NM_021800
NM_201262
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr7_+_75769927
|
0.382
|
|
HSPB1
|
heat shock 27kDa protein 1
|
|
chr17_-_44400952
|
0.382
|
NM_004123
|
GIP
|
gastric inhibitory polypeptide
|
|
chr11_+_18390421
|
0.381
|
NM_002301
NM_017448
|
LDHC
|
lactate dehydrogenase C
|
|
chr1_-_56817717
|
0.380
|
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr14_+_57964148
|
0.378
|
|
|
|
|
chr10_-_14919973
|
0.378
|
NM_001029954
|
CDNF
|
cerebral dopamine neurotrophic factor
|
|
chr6_+_27914301
|
0.378
|
NM_003520
|
HIST1H2BN
|
histone cluster 1, H2bn
|
|
chr5_+_137447479
|
0.375
|
NM_058244
|
WNT8A
|
wingless-type MMTV integration site family, member 8A
|
|
chr10_+_96688399
|
0.375
|
NM_000771
|
CYP2C9
|
cytochrome P450, family 2, subfamily C, polypeptide 9
|
|
chr7_+_93862137
|
0.372
|
|
COL1A2
PLAG1
|
collagen, type I, alpha 2
pleiomorphic adenoma gene 1
|
|
chr8_-_122722674
|
0.371
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr19_-_20540380
|
0.370
|
NM_001159293
|
ZNF737
|
zinc finger protein 737
|
|
chr12_+_15590552
|
0.370
|
NM_030668
NM_030669
NM_030670
NM_030671
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr1_+_25629968
|
0.369
|
NM_018202
|
TMEM57
|
transmembrane protein 57
|
|
chr5_-_42847716
|
0.368
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr1_-_116112684
|
0.364
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr2_+_102744921
|
0.363
|
NM_144632
|
TMEM182
|
transmembrane protein 182
|
|
chr4_-_56153135
|
0.361
|
NM_152401
|
PDCL2
|
phosducin-like 2
|
|
chr17_-_59437782
|
0.359
|
NM_000873
|
ICAM2
|
intercellular adhesion molecule 2
|
|
chr3_+_190147633
|
0.358
|
|
TPRG1
|
tumor protein p63 regulated 1
|
|
chrX_+_131179886
|
0.358
|
|
|
|
|
chr6_+_12825818
|
0.358
|
NM_030948
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr5_-_58688428
|
0.358
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr17_-_10362583
|
0.356
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr2_-_197383120
|
0.354
|
NM_213608
|
C2orf66
|
chromosome 2 open reading frame 66
|
|
chr2_+_38746555
|
0.354
|
NM_138801
|
GALM
|
galactose mutarotase (aldose 1-epimerase)
|
|
chr6_-_48186868
|
0.352
|
|
C6orf138
|
chromosome 6 open reading frame 138
|
|
chr1_+_239882202
|
0.352
|
NM_144625
|
WDR64
|
WD repeat domain 64
|
|
chr14_+_23670514
|
0.349
|
NM_203402
|
FITM1
|
fat storage-inducing transmembrane protein 1
|
|
chr2_-_148494732
|
0.347
|
NM_001190882
NM_001190879
NM_002552
NM_001190881
NM_181741
|
ORC4
|
origin recognition complex, subunit 4
|
|
chr1_-_116112853
|
0.346
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr4_-_95482994
|
0.344
|
NM_014485
|
HPGDS
|
hematopoietic prostaglandin D synthase
|
|
chr8_+_76614757
|
0.344
|
NM_004133
|
HNF4G
|
hepatocyte nuclear factor 4, gamma
|
|
chr1_-_147665783
|
0.344
|
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr3_+_120982020
|
0.343
|
NM_003889
NM_033013
|
NR1I2
|
nuclear receptor subfamily 1, group I, member 2
|
|
chr4_+_86918870
|
0.342
|
NM_001042669
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr1_-_148959945
|
0.341
|
NM_032132
|
HORMAD1
|
HORMA domain containing 1
|
|
chr4_+_146820805
|
0.341
|
NM_001080531
|
C4orf51
|
chromosome 4 open reading frame 51
|
|
chr3_+_35658852
|
0.339
|
NM_016300
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr4_-_186970366
|
0.338
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr3_-_150533948
|
0.337
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr19_+_12724406
|
0.336
|
NM_017682
|
BEST2
|
bestrophin 2
|
|
chr6_-_27548438
|
0.335
|
NM_007149
|
ZNF184
|
zinc finger protein 184
|
|
chrX_+_70438352
|
0.335
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chr1_-_156923111
|
0.333
|
NM_003126
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2)
|
|
chr17_+_5913149
|
0.333
|
|
WSCD1
|
WSC domain containing 1
|
|
chr12_-_120591938
|
0.329
|
NM_173855
|
MORN3
|
MORN repeat containing 3
|
|
chr4_-_187114407
|
0.328
|
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr5_+_34875025
|
0.327
|
NM_144725
|
TTC23L
|
tetratricopeptide repeat domain 23-like
|
|
chr10_+_80569464
|
0.326
|
|
ZMIZ1
|
zinc finger, MIZ-type containing 1
|
|
chr1_+_154605626
|
0.325
|
NM_020407
|
RHBG
|
Rh family, B glycoprotein (gene/pseudogene)
|
|
chr7_+_94953148
|
0.325
|
NM_016116
NM_145872
|
ASB4
|
ankyrin repeat and SOCS box containing 4
|